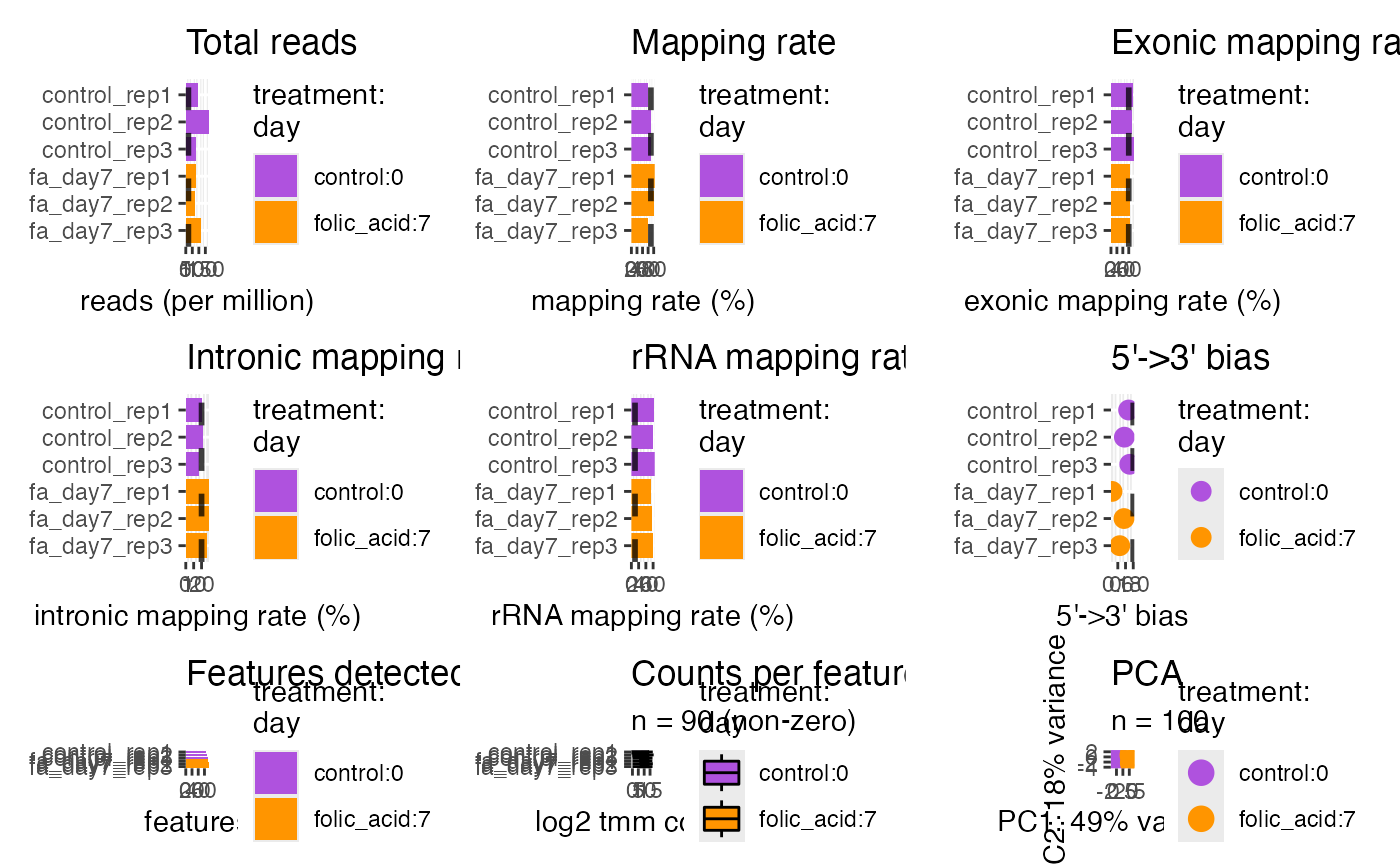
Quality control
Examples
data(bcb)
## bcbioRNASeq ====
plotQc(bcb)
#> ℹ Using "tmm" counts.
#> → Applying trimmed mean of M-values (TMM) normalization.
#> ℹ Filtered zero count rows and columns:
#> - 90 / 100 rows (90%)
#> - 6 / 6 columns (100%)
#> → Applying `log2(x + 1L)` transformation.
#> ℹ Using "vst" counts.