Plot feature (gene) detection saturation
Source:R/AllGenerics.R, R/plotFeatureSaturation-methods.R
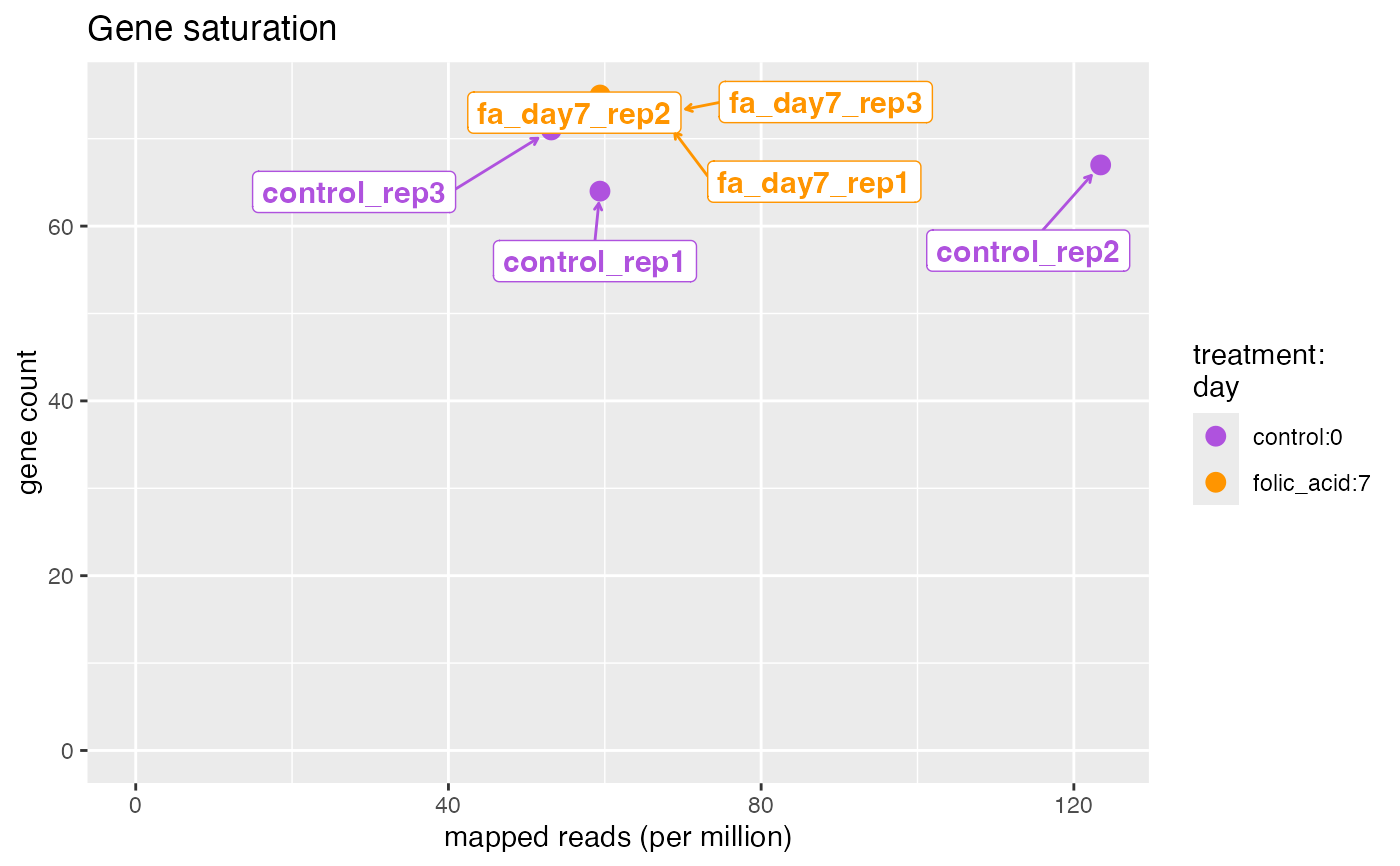
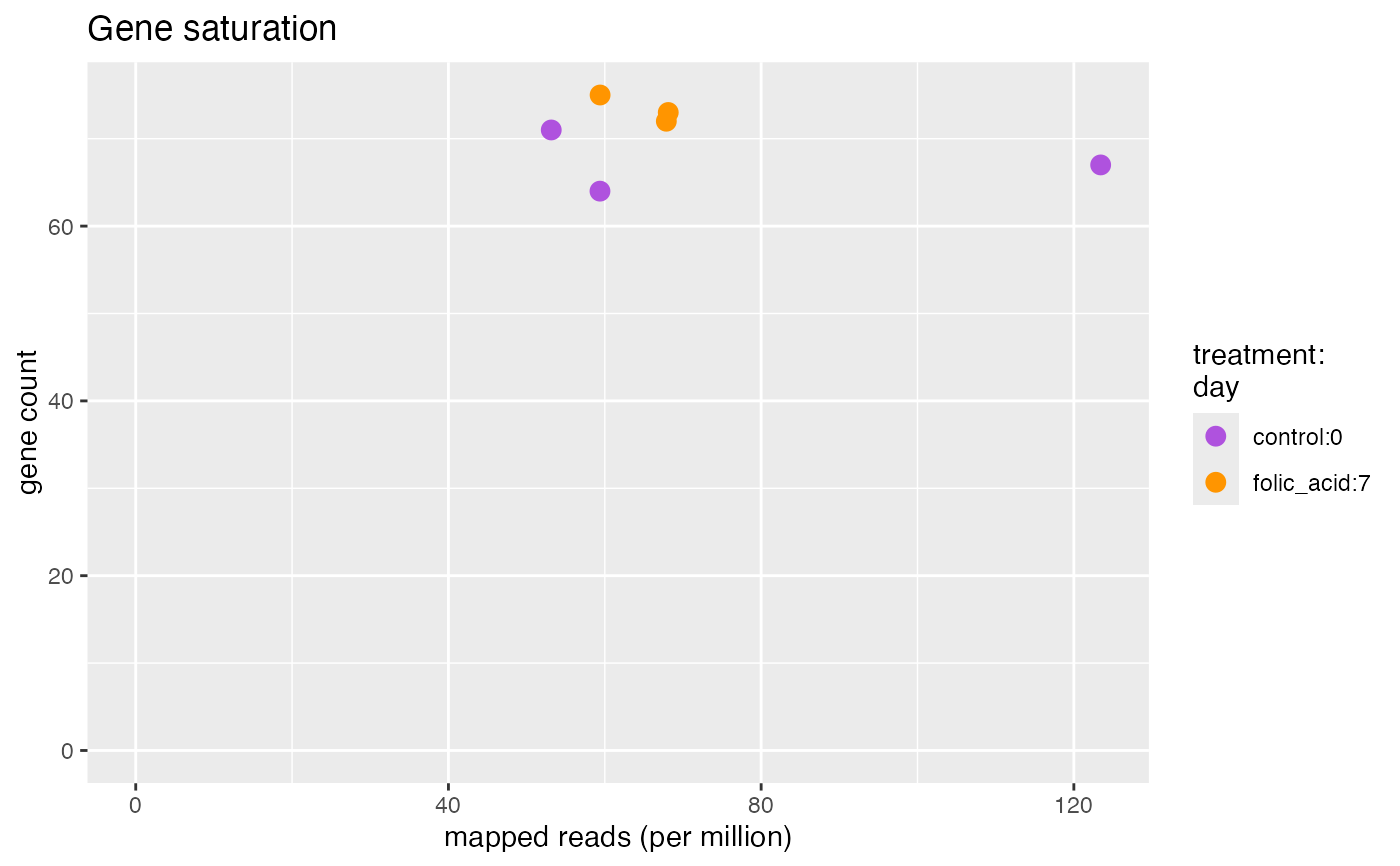
plotFeatureSaturation.RdWe should observe a linear trend in the number of genes detected with the number of mapped reads, which indicates that the sample input was not overloaded.
Usage
plotFeatureSaturation(object, ...)
# S4 method for bcbioRNASeq
plotFeatureSaturation(
object,
interestingGroups = NULL,
minCounts = 1L,
perMillion = TRUE,
trendline = FALSE,
label = getOption(x = "acid.label", default = FALSE),
labels = list(title = "Gene saturation", subtitle = NULL, x = "mapped reads", y =
"gene count")
)Arguments
- object
Object.
- interestingGroups
character. Groups of interest to use for visualization. Corresponds to factors describing the columns of the object.- minCounts
integer(1). Minimum number of counts per feature (i.e. gene).- perMillion
logical(1). Display as counts per million.- trendline
logical(1). Include trendline on plot.- label
logical(1). Superimpose sample text labels on the plot.- labels
list. ggplot2 labels. Seeggplot2::labs()for details.- ...
Additional arguments.
Examples
data(bcb)
## bcbioRNASeq ====
plotFeatureSaturation(bcb, label = FALSE)
 plotFeatureSaturation(bcb, label = TRUE)
plotFeatureSaturation(bcb, label = TRUE)