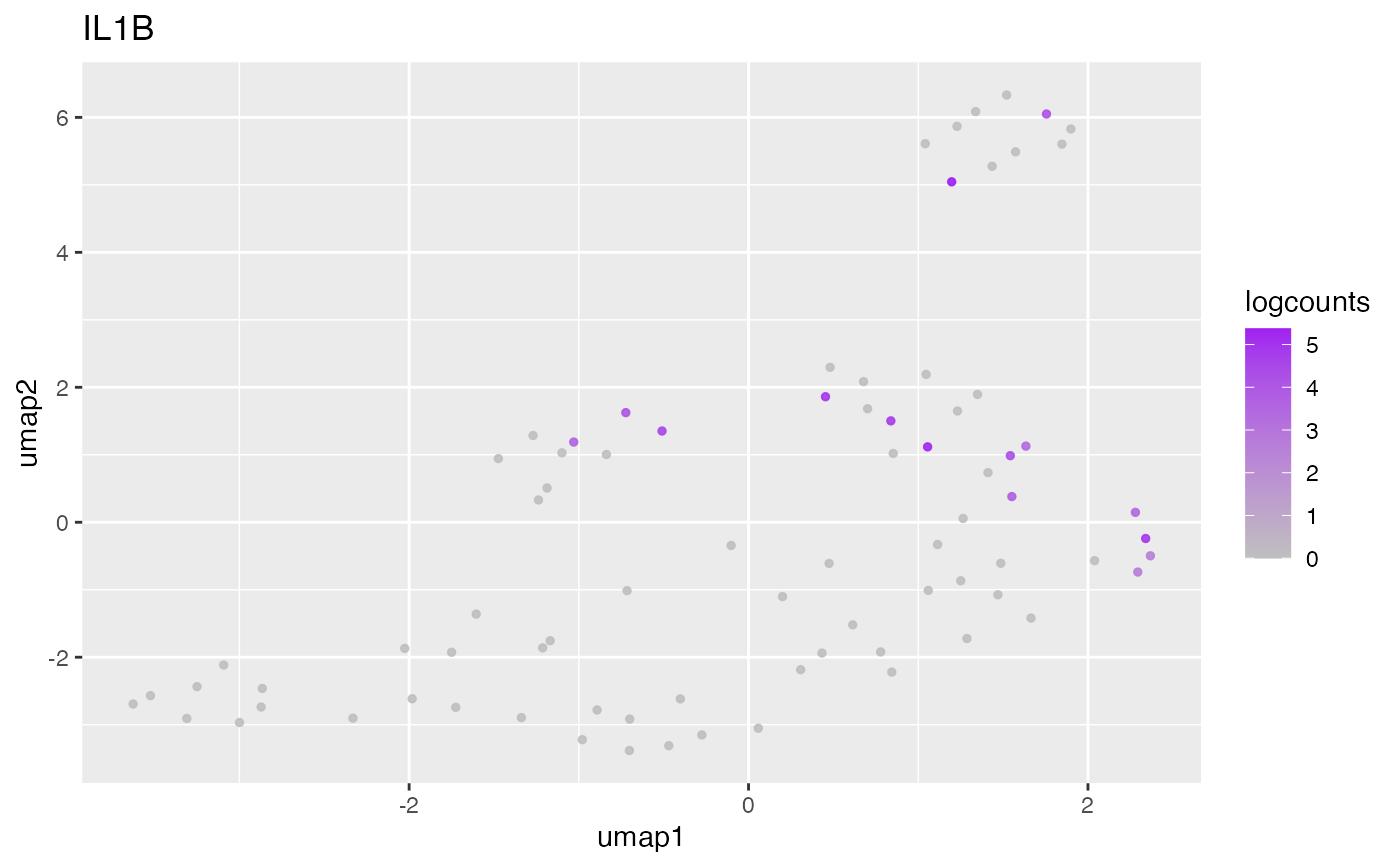
Plot known markers
Usage
plotKnownMarkers(object, markers, ...)
# S4 method for class 'SingleCellExperiment,KnownMarkers'
plotKnownMarkers(object, markers, reduction = "UMAP", headerLevel = 2L, ...)Arguments
- object
Object.
- markers
Object.
- reduction
vector(1). Dimension reduction name or index position.- headerLevel
integer(1)(1-7). Markdown header level.- ...
Passthrough arguments to
plotMarker().
Examples
data(
KnownMarkers,
SingleCellExperiment_Seurat,
package = "AcidTest"
)
## SingleCellExperiment ====
object <- SingleCellExperiment_Seurat
markers <- KnownMarkers
plotKnownMarkers(
object = object,
markers = markers,
reduction = "UMAP"
)
#>
#>
#> ## Dendritic Cell {.tabset}
#>
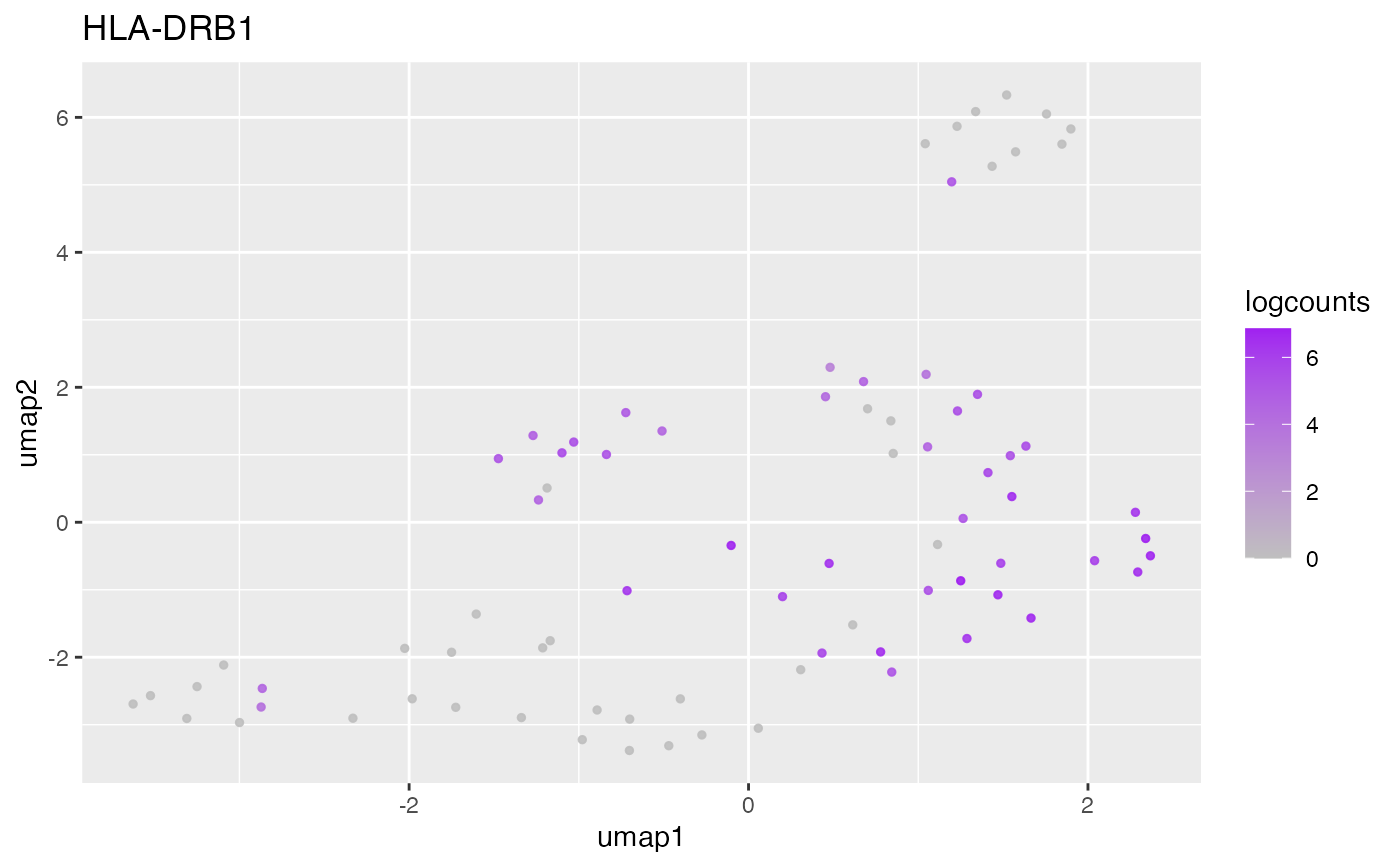
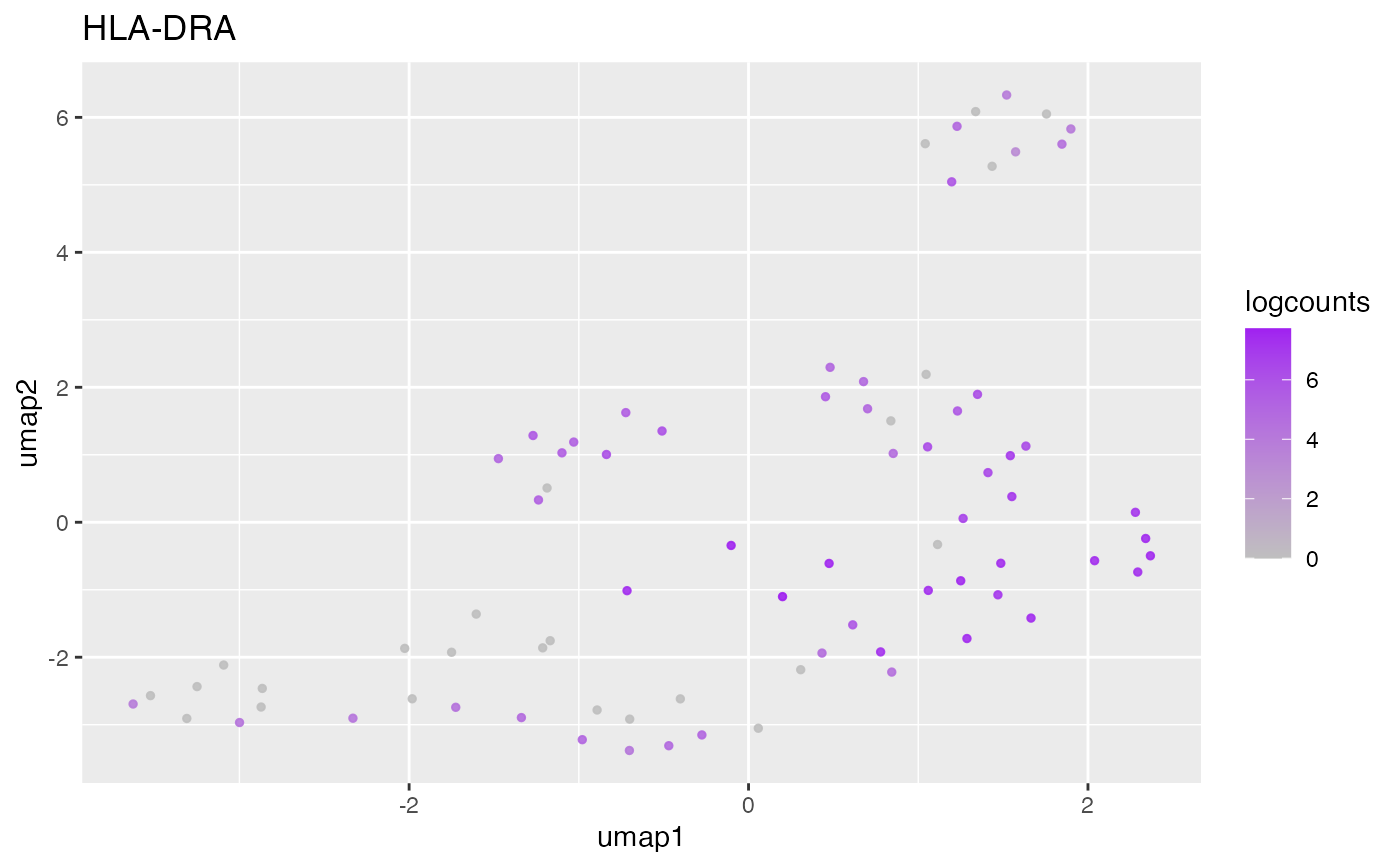
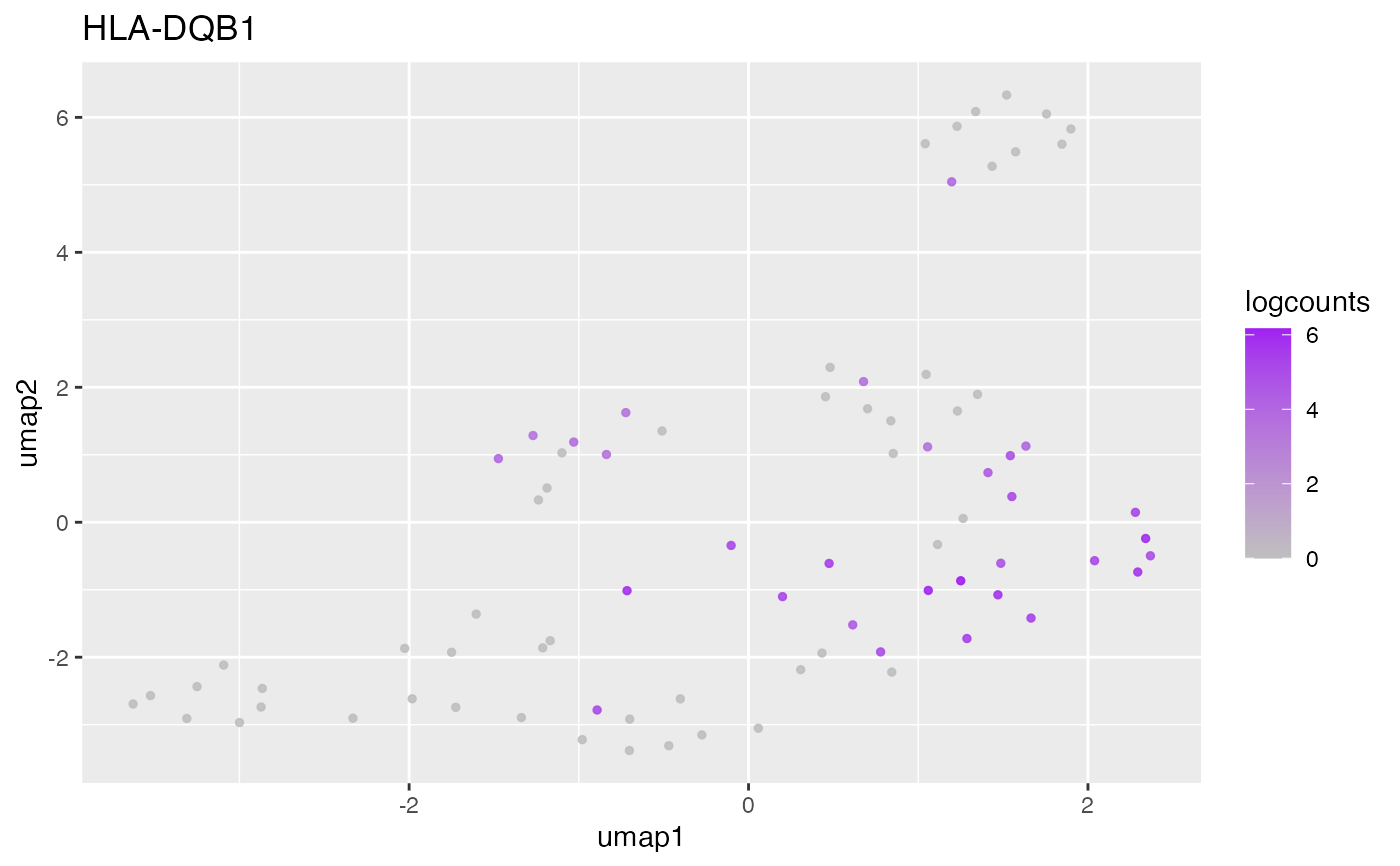
 #>
#>
#> ## Macrophage {.tabset}
#>
#>
#>
#> ## Macrophage {.tabset}
#>